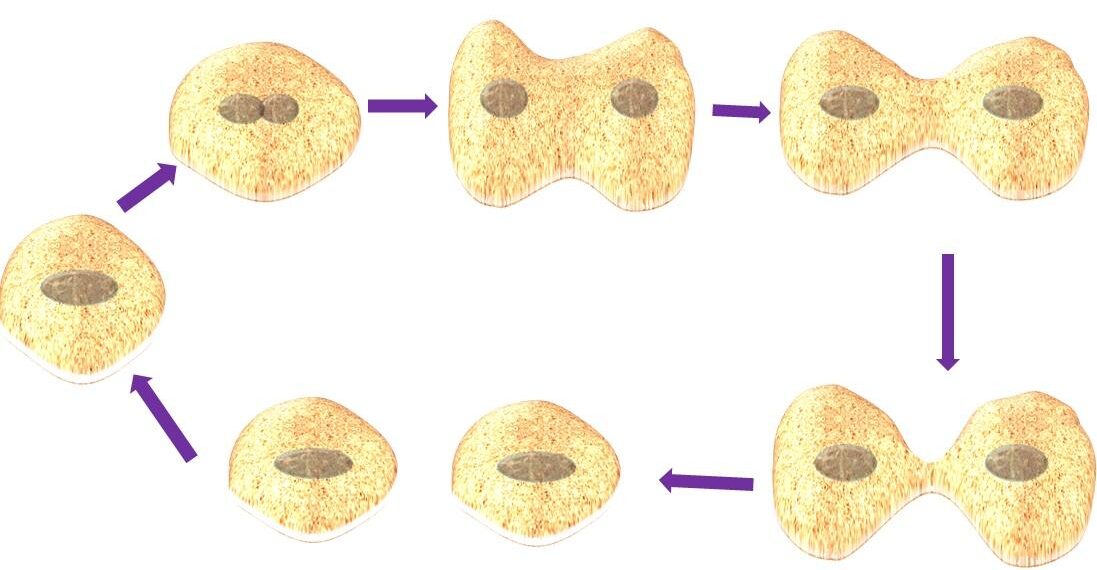
Prokaryotic DNA replication is an anabolic process by which prokaryotes duplicate their genomic DNA to transfer it to their daughter cells.
DNA replication in prokaryotes especially in E. coli (model microorganism) is well studied.
DNA replication of prokaryotes (e.g. E. coli)
DNA replication is not a random process, so replication starts at a particular site in the chromosome.
The genomic DNA of prokaryotes is organized into a single replicon. Replicon is the unit of replication, that contains 4.6 Mbp size and it indicates an entire DNA that can replicate as a single unit.
Each replicon contains a single origin called the origin of replication or chromosomal origin (ori-C). Ori-C is a 244 base pair sequence that contains several types of DNA replicative sequences.
The unit (replicon) of DNA is replicated from ori-C by bidirectionally or uni-directionally to the terminus site. The total genome of E. coli is a single replicon and its size is approximately 4.6 Mbp, hence the whole genome in a bacterium replicates as a single unit.
Proteins required for prokaryotic DNA replication
DNA replication is a complex process that occurs in three steps including initiation, elongation, and termination. These steps require several enzymes and proteins. The studies found that more than 20 different proteins play a central role in prokaryotic DNA replication. These proteins have been classified on the basis of their biochemical functions in different stages of prokaryotic DNA replication.
1) Proteins required for replication initiation
DnaA (initiator)
Single strand binding protein (SSB)
DnaB (helicase)
DnaC (helicase loader)
DnaG (primase)
2) Proteins involved in DNA chain elongation and ligation of Okazaki fragments:
DNA polymerase (adds the nucleotides)
SSB (stabilizes the single strands)
DNA gyrase (unwinds the strands)
DNA ligase (joins the Okazaki fragments)
3) Proteins required for replication termination and separation of daughter DNA molecules
Tus protein (terminus binding protein)
DNA topoisomerase (separates daughter molecules)
The major repetitive sequences present in ori-c are:
1) Four 9 mer sequences:- These are the binding sites for the replication initiation protein such as Dna-A.
2) 3.13 mer sequences:- These are AT-rich sequences, which undergo initial melting during the replication initiation.
3) Several GATC sequences:- These sequences are palindromes, that read both backward as forwards same. The adenine base in the GATC sequence can be methylated and methylation status determines the occurrence of replication initiation. If the GATC sequence is methylated in both the strands then the initiation protein can bind to the DNA and start the replication initiation. However, if GATC is hemimethylated then the initiation protein cannot bind to the DNA, thereby replication will not be initiated.
Mechanism of DNA replication in prokaryotes in several steps
Initiation
Step 1: The replication initiation begins with the binding of Dna-A to five 9 mer sequences present in the oriC region. The binding of Dna-A to 9 mer sequence will be initially very week and will have low binding efficiency. However, binding of the first Dna-A protein exhibits cooperativity, which resulted in increasing the binding efficiency of subsequent Dna-A proteins to 9 mere sequences.
Step 2. The binding of Dna-A followed by binding of Histone-like protein called Hu proteins. The binding of Dna-A and Hu protein at 9 mer sequence causes a conformational change in adjacent 13 mere sequence leading to the localized melting of DNA strand separation. This process uses ATP and forms an open complex.
Step 3. The localized unwinding of DNA in the 13 mer sequence promotes the binding of helix destabilization proteins SSBs (single-strand binding proteins).
Step 4. DNA helicase, Dna-B (hexamer), is loaded onto the 13 mere sequence by the helicase loader (Dna-C). Hexameric identical subunits of DNA helicase will clamp around each of the two single strands formed as an open complex between DnaA and oriC. This process requires ATP. As the unwinding of DNA occurs in the presence of DNA helicase, the initiation protein DnaA and Hu proteins are gradually removed from 9 mer sequences.
Step 5. DNA polymerase can only elongate existing primer strands of DNA or RNA. Hence. the unwinding of DNA by DNA helicase facilitates the binding of the primase enzyme (Dna-G) to the helicase and forms the primosome complex. Then, DNA primase synthesizes the first primer for the leading strand.
Elongation
Step 6: The primer formation then promotes loading of the sliding clamp by the clamp loader (γ-complex). The core DNA polymerase is then added to the sliding clamp. Once the core polymerase is loaded then it begins primer extension.
Step 7: When the primer of the leading strand is extended by few nucleotides then the first primer for the lagging strand is synthesized. The primer synthesizing activity will have to be repeated several times during the formation of the lagging strand, which is synthesized in the form of Okazaki fragments, ranging in the size from 1000 to 2000 nucleotides.
In order to facilitate the same rate of replication in both leading and lagging strands, the lagging strand template makes loop formation, so that both the strands will be replicated with the same duration. The usual rate of replication by DNA polymerase-III is 200 to 1000 nucleotides incorporation per second, under optimal conditions DNA polymerase can add 15000 nucleotides per minute.
Termination
Step 8: The elongation phase continues until the polymerases reach the termination sequence, each DNA strand contains a Ter sequence opposite to the origin sequence, the two Ter sequences in E. coli are Ter EDA and Ter BCF. Ter sequence is a binding site for Tus sequence.
Step 9: When the Tus protein binds to the Ter sequence then it will not allow the DNA polymerase to move at the Ter region and therefore replication will be terminated.
Step 10: In the case of the lagging strand, the Okazaki fragments can join together by DNA ligase to complete the replication process.
Key points
- The time required for replication of whole-genome and doubling time in E. coli is virtually constant at 37ºC.
- The time required for the replication of the chromosome is 40 min and 20 min for doubling the cell.
- To continue the doubling of cells, the organism should develop its capacity to initiate the second round of replication before the previous round has terminated.
- Because of this advanced initiation process, on a single chromosome, approximately six replication forks may be developed, hence, this process is called Dichotomous replication.
Frequently asked questions
What is required for DNA replication to begin in a prokaryotic cell?
Answer: DnaA (initiator)
Which of the following components required for prokaryotic DNA replication?
1. Dna B, Dna C and Dna G
2. EF-Tu and EF-G,
3. DNA pol-α DNA pol-δ and DNA pol-ε
Answer: DnaB (helicase), DnaC (helicase loader), and DnaG (primase)
Does DNA replication in prokaryotes require Okazaki fragments?
Answer: Yes
Explanation: Prokaryotic cells also contain Okazaki fragments as eukaryotic cells. However, they are a little longer than those of eukaryotic cells. Okazaki fragments of prokaryotic cells (e.g. E. coli) are 1000-2000 nucleotides long, while the fragments in a eukaryotic cell will be 100 to 200 nucleotides long.