Ubiquitination definition
Ubiquitination is a post-translational modification process that plays a major role in protein degradation. The ubiquitin-proteasome system is involved in the process called ubiquitination, ubiquitylation, or molecular “kiss of death”. There are two main protein degrading systems that are found in eukaryotic cells, including the lysosome proteolytic system and the proteasome proteolytic system.
The proteins present in the cells are under the surveillance of cellular degradative machinery. If any unfavorable changes occur in the protein structure, then the protein will be taken by this machinery to protect the cells from malfunction.
Ubiquitination key points
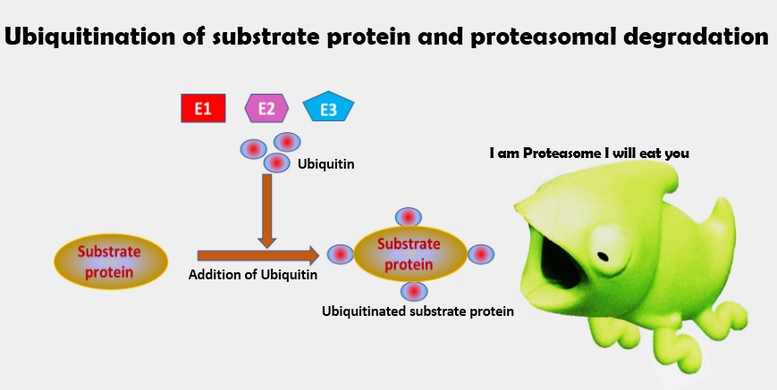
- Ubiquitin proteins are covalently attached to the substrate proteins (damaged proteins) to initiate the degradation; hence, this process is termed ubiquitination.
- Prokaryotes have no molecules that are functionally equivalent to ubiquitin. However, structurally ubiquitin-related proteins such as Ubiquitin Bacterial (UBact) have been found in bioinformatics studies.
- The monomer of ubiquitin contains 76 amino acids that are folded into a compact, globular, tightly hydrogen-bonded structure with a terminal signature diglycine sequence. However, in some cases, diglycine-modified lysine is used as a signature of ubiquitination.
- It also recognizes the specific site of modification.
- Ubiquitin protein is initially synthesized in a precursor form, thereby generating the signature Gly–Gly sequence, which must be processed by deubiquitylating enzymes.
- Ubiquitination is an active enzymatic process that requires energy released from ATP hydrolysis.
- The process is highly specific and degrades only those proteins that are marked by ubiquitination.
- In ubiquitination, the lysin residue in the damaged protein will be conjugated with the ubiquitin protein.
- The ubiquitination process is highly specific; hence, the cytosolic degradation machinery recognizes only ubiquitinated proteins (this process cannot randomly degrade other proteins).
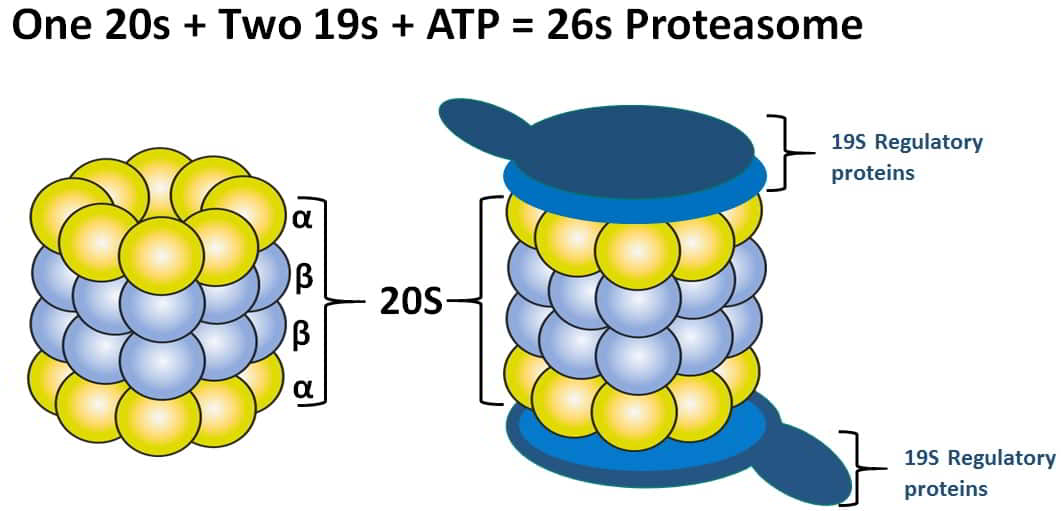
- Cytosolic protein degradation is carried out by a 26S proteasomal complex that contains core 21S proteasomes.
- Ubiquitin is a low molecular weight heat shock protein that finds and marks substrate proteins for proteasomal degradation.
- This process requires three more proteins called E1, E2, and E3.
- The E1 protein is a ubiquitin activase that activates ubiquitin.
- The E2 protein is a ubiquitin conjugase that binds to activated ubiquitin and facilitates the binding of ubiquitin to the substrate protein.
- E3 is a ubiquitin ligase that recognizes the protein to be degraded or to be ubiquitinated.
- The eukaryotic cell contains different types of E3 proteins that can target different parameters of proteins for degradation. For instance, some E3 proteins recognize the N-terminal, some E3 proteins identify the PEST sequence, some recognize the oxidized-side chains, and others recognize misfolding.
- During ubiquitination, the activated ubiquitin combines with the Lys residue present in the substrate protein, which is catalyzed by E3.
- The ubiquitinated protein is transferred to the 26S proteasomal complex, where the ubiquitin proteins are separated from substrate proteins and recycled for further substrate protein activation. Now the ubiquitin protein has entered the proteasomal core and will be degraded into octapeptides (small fragments).
Ubiquitination definition
Ubiquitination is a post-translational modification process that plays a major role in protein degradation. The ubiquitin-proteasome system is involved in the process called ubiquitination, ubiquitylation, or molecular “kiss of death”. There are two main protein degrading systems that are found in eukaryotic cells, including the lysosome proteolytic system and the proteasome proteolytic system.
The proteins present in the cells are under the surveillance of cellular degradative machinery. If any unfavorable changes occur in the protein structure, then the protein will be taken by this machinery to protect the cells from malfunction.
Ubiquitination key points
- Ubiquitin proteins are covalently attached to the substrate proteins (damaged proteins) to initiate the degradation; hence, this process is termed ubiquitination.
- Prokaryotes have no molecules that are functionally equivalent to ubiquitin. However, structurally ubiquitin-related proteins such as Ubiquitin Bacterial (UBact) have been found in bioinformatics studies.
- The monomer of ubiquitin contains 76 amino acids that are folded into a compact, globular, tightly hydrogen-bonded structure with a terminal signature diglycine sequence. However, in some cases, diglycine-modified lysine is used as a signature of ubiquitination.
- It also recognizes the specific site of modification.
- Ubiquitin protein is initially synthesized in a precursor form, thereby generating the signature Gly–Gly sequence, which must be processed by deubiquitylating enzymes.
- Ubiquitination is an active enzymatic process that requires energy released from ATP hydrolysis.
- The process is highly specific and degrades only those proteins that are marked by ubiquitination.
- In ubiquitination, the lysin residue in the damaged protein will be conjugated with the ubiquitin protein.
- The ubiquitination process is highly specific; hence, the cytosolic degradation machinery recognizes only ubiquitinated proteins (this process cannot randomly degrade other proteins).
- Cytosolic protein degradation is carried out by a 26S proteasomal complex that contains core 21S proteasomes.
- Ubiquitin is a low molecular weight heat shock protein that finds and marks substrate proteins for proteasomal degradation.
- This process requires three more proteins called E1, E2, and E3.
- The E1 protein is a ubiquitin activase that activates ubiquitin.
- The E2 protein is a ubiquitin conjugase that binds to activated ubiquitin and facilitates the binding of ubiquitin to the substrate protein.
- E3 is a ubiquitin ligase that recognizes the protein to be degraded or to be ubiquitinated.
- The eukaryotic cell contains different types of E3 proteins that can target different parameters of proteins for degradation. For instance, some E3 proteins recognize the N-terminal, some E3 proteins identify the PEST sequence, some recognize the oxidized-side chains, and others recognize misfolding.
- During ubiquitination, the activated ubiquitin combines with the Lys residue present in the substrate protein, which is catalyzed by E3.
- The ubiquitinated protein is transferred to the 26S proteasomal complex, where the ubiquitin proteins are separated from substrate proteins and recycled for further substrate protein activation. Now the ubiquitin protein has entered the proteasomal core and will be degraded into octapeptides (small fragments).
What does ubiquitin do in the cell?
The major functions of ubiquitin in the cell include
- Ubiquitin is involved in the MHC class I-restricted antigen processing of ER-targeted proteins and also plays a major role in the processing of cytosolic proteins.
- Ubiquitination is also involved in apoptosis.
- It plays a major role in the biosynthesis of cellular organelles
- Ubiquitin regulates cell cycle and cell division and is also responsible for several major cell cycle transitions via selective degradation of cyclin-dependent kinase inhibitors, cyclins, and anaphase inhibitors.
- It regulates DNA transcription and DNA repair.
- UPS is involved in cell differentiation and development.
- Participates in neural and muscular degeneration.
- Regulates dynamic cellular mechanisms such as endocytosis, proteolysis, and immune signaling.
- Involved morphogenesis of neural networks.
- UPS is involved in inflammation and immune responses.
- This protein plays a major role in the modulation of cell surface receptors, ion channels, and secretory pathways.
- Respond to stress and extracellular modulators.
- UPS is involved in ribosomal biogenesis.
- Regulates viral infection, however, some viruses like influenza A virus (IAV) seize ubiquitination and ubiquitin-like modifications to establish viral infection.
Ubiquitination and other proteolytic mechanisms
Ubiquitination is a proteasomal complex-mediated proteolytic process that uses various ubiquitin proteins to recruit the substrate protein for degradation.
Protein degradation is a required process in all eukaryotic cells. Hence, the cells own two types of proteolytic systems, including lysosomal degradation and cytosolic degradation (proteasomal-mediated degradation).
These two mechanisms are mainly involved in the degradation of aged or damaged proteins to keep the cells clean and healthy. Within these two mechanisms, lysosomes-mediated (membrane-bound organelles) protein degradation play a pivotal role in the recycling of cellular waste materials. Lysosomes contain an assortment of acid hydrolases and many essential enzymes.
Mechanism of Ubiquitination (ubiquitylation of substrate protein)
Ubiquitin-mediated protein degradation is a well-established mechanism to send damaged proteins to proteasomes by their modification with chains of a protein called ubiquitin.
In eukaryotes, this is a highly conserved protein that contains 76 amino acids and is usually bound with the substrate by an isopeptide bond, which forms between a lysine residue of the substrate and the C-terminal glycine of ubiquitin.
Sequential steps of the ubiquitination process
*Substrate protein will be activated in three steps before going to proteasomal degradation
STEP: 1 In most organisms, a single E1 (ubiquitin-activating enzyme) binds covalently with free ubiquitin-protein via the C-terminal residue of ubiquitin using energy from ATP.
STEP: 2 The activated ubiquitin is subsequently transferred to a cysteine on a ubiquitin-conjugating enzyme 2 (E2).
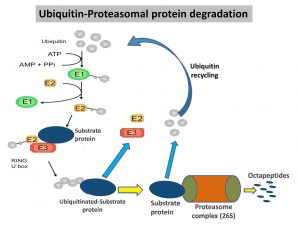
STEP: 3 Then, the ubiquitin-protein ligases (E3) transfer the activated ubiquitin from E2 to the lysin residue of the final target protein (substrate), forming an isopeptide bond.
Finally, this ubiquitinated substrate protein will be proteolytically degraded into small peptide fragments by the proteasome, which is an ATP-dependent process. Then, the released fragments are subsequently engulfed by lysosomes for further degradation (complete lysis).
Frequently asked questions related to Ubiquitin
1. What is ubiquitin, and what role does it play in tagging proteins for degradation? or what is the purpose of ubiquitin? or what is the role of ubiquitin?
Answer: Ubiquitin is a small regulatory polypeptide that is found in all eukaryotes. The protein plays a crucial role in the degradation of unwanted proteins. In general, all proteins of the cells are under the surveillance of cellular degradation machinery (ubiquitination). If there are any unfavorable changes in the protein structure, then the protein will be degraded by ubiquitination to protect the cells from malfunction.
2. Ubiquitin tags proteins for what purpose?
Answer: Ubiquitin proteins attach to substrate proteins (mutant proteins) to initiate their degradation using ubiquitination machinery.
3. Which of the following is most likely to have a small protein called ubiquitin attached to it?
A. The mRNA that is leaving the nucleus to be translated.
B. a cell surface protein that requires transport from the ER.
C. a cyclin protein that is usually required in the G1 phase, now that the cell is in the G2 phase
D. a regulatory protein that needs a sugar molecule to be attached
E. an mRNA produced by an egg cell that will be retained until after fertilization.
Answer: C
Read the subsequent process that occurs in proteasomes.
References
- Alina Rudnicka and Yohei Yamauchi. Ubiquitin in Influenza Virus Entry and Innate Immunity. Viruses. 2016 Oct; 8(10): 293. Published online 2016 Oct 24. doi: [10.3390/v8100293]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004 Nov 29;1695(1-3):55-72.
- The ubiquitin-proteasome pathway. Annual Reviews Collection [Internet].
- de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998 Apr 17;273(16):9734-43.
- Ubiquitin B [ Mus musculus (house mouse).
- Kravtsova-Ivantsiv Y, Ciechanover A. Ubiquitination and degradation of proteins. Methods Mol Biol. 2011;753:335-57. doi: 10.1007/978-1-61779-148-2_23.