Marker enzymes definition: Marker enzymes are biomarkers, which are not frequent but limited to and synthesized by specific cell organelles. Hence, the marker enzymes are used to find and isolate the specific organelle from complex cellular compartments.
The well-organized eukaryotic organisms, such as animals and plants are composed of millions of cells. Even though each cell type of the body, derived from a single fertilized cell during development and contains a complete set of chromosomes similar to other cells of that body, under the microscope they may appear different from others.
All cells are genetically the same but functionally they are differentiated in various tissues, such as skin, muscle, nerves, bone, blood, etc. These specialized tissues express only specific subsets of the whole genome resulting in exploiting the specialized metabolic pathways and keeping others biologically inactive.
If we jump into the cytoplasm of the cells, all cells of the body contain the same set of organelles irrespective of their location and function. These cellular organelles present in specified regions within the cell and involve various metabolic functions.
The development and organization of these internal cellular compartments (organelles) in the proper position and range within the cell allow cells to get differentiation within the tissues and organs.
In laboratory conditions, there is a generalized analogy, which uses marker enzymes as experimental tools to separate the particular organelle from others. In addition, using these marker enzymes, the cellular abnormalities within the specified tissue can be screened.
Marker enzymes of organelles and their function
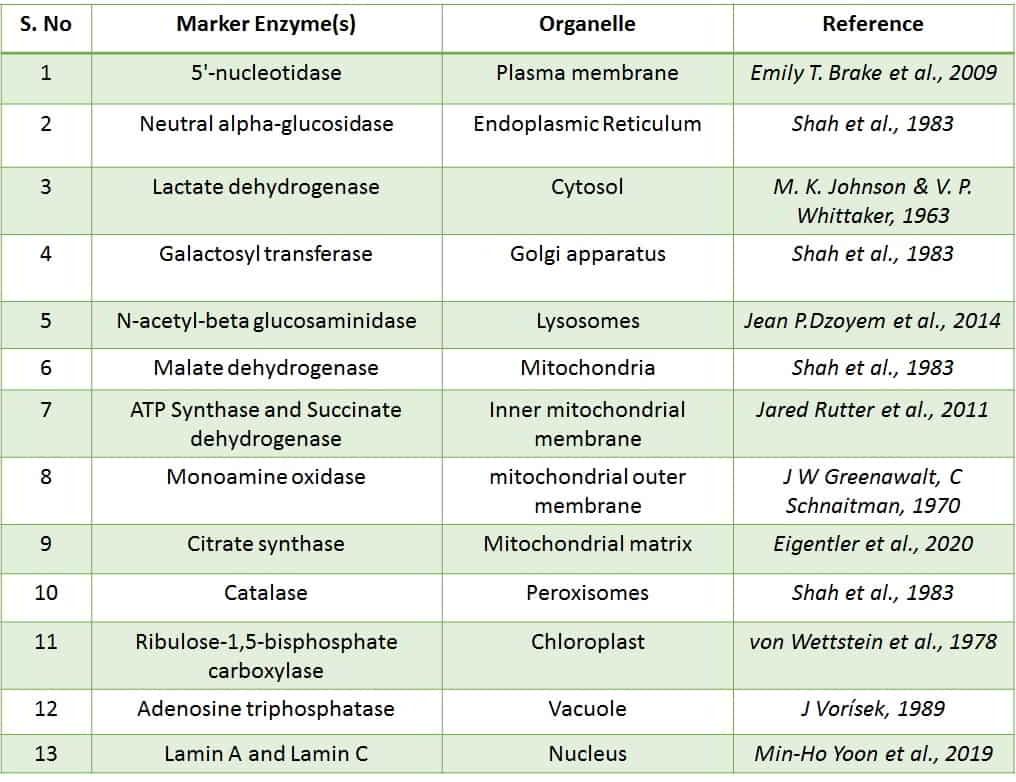
Marker enzymes are those enzymes that are confined to a specific organelle and they help in differentiating an organelle from others.
Organelles and their marker enzymes
The cytosol is an intracellular fluid, called a cytoplasmic matrix, which is made up of water and water-soluble inorganic and organic compounds. It also contains suspended insoluble particles like ribosomes, which play a central role in protein translation.
- Lactate dehydrogenase (LDH) is a marker enzyme of cytosol.
- 5′-nucleotidase is a marker enzyme of Plasma membrane (PM).
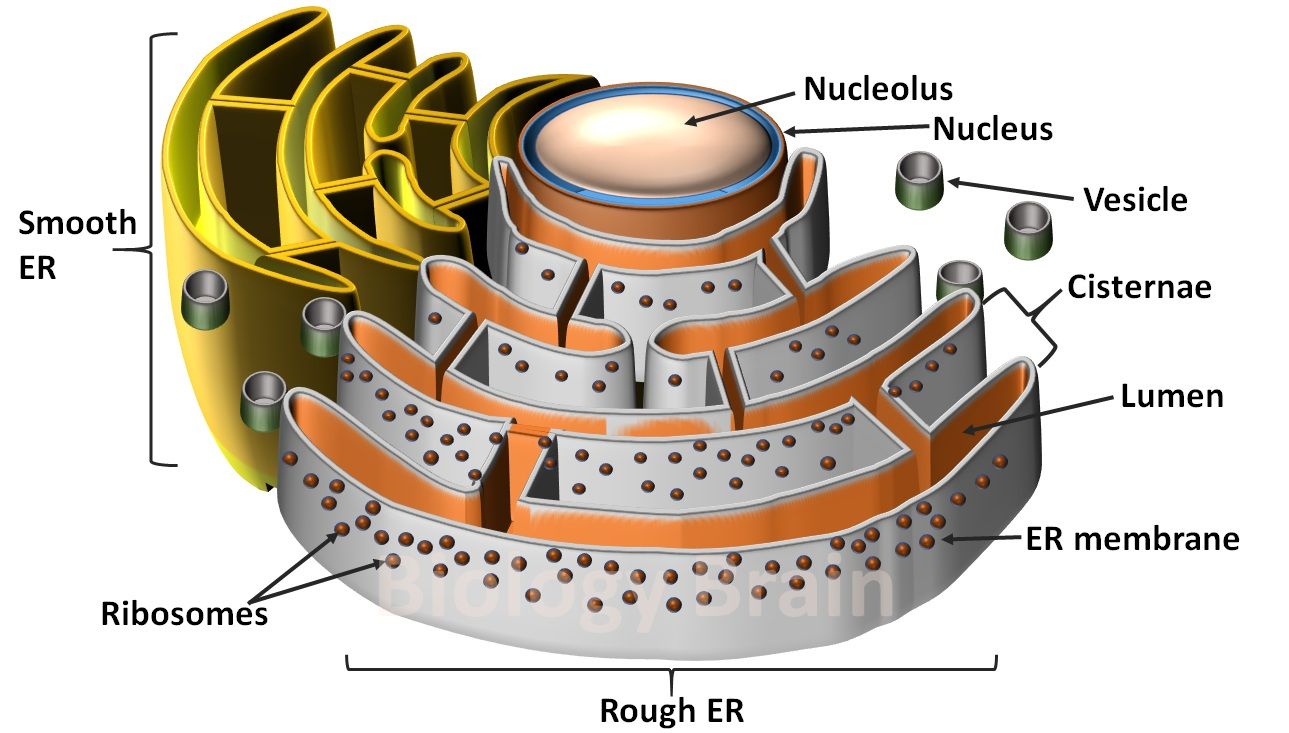
- Neutral alpha-glucosidase is a marker enzyme of the Endoplasmic Reticulum (ER).
- Galactosyl transferase is a marker enzyme of the Golgi apparatus (GC).
- N-acetyl-beta glucosaminidase is a marker enzyme of Lysosomes.
- Malate dehydrogenase is a marker enzyme of Mitochondria.
- ATP Synthase and Succinate dehydrogenase are marker enzymes of the Inner mitochondrial membrane.
- Monoamine oxidase is a marker enzyme of the mitochondrial outer membrane.
- Citrate synthase is a marker enzyme of the Mitochondrial matrix.
- Catalase is a marker enzyme of Peroxisomes.
- Ribulose-1,5-bisphosphate carboxylase is an enzyme of Chloroplast.
- Adenosine triphosphatase (ATPase) is a marker enzyme of Vacuole.
- Lamin A and Lamin C are the marker proteins of the Nucleus.
Enzyme markers for organelle separation by centrifugation
If we get the human lymphocytes to characterize their cellular organelles using their marker enzymes, first we should lyse the cell membrane with erythrocyte hypotonic lysis buffer. Then the obtained homogenates of mixed lymphocytes have to be subjected to analytical sub-cellular fractionation using sucrose gradient centrifugation. The centrifugation process separates the cellular compartments in different gradients based on their weight and density in the suspension buffer. According to their density, we can separate the layers containing specific organelles from the centrifuge tube, and finally, the isolated organelles can be identified using their marker enzymes or proteins.
Data source
- Shah T, Webster AD, Peters TJ. Enzyme analysis and subcellular fractionation of human peripheral blood lymphocytes with special reference to the localization of putative plasma membrane enzymes. Cell Biochem Funct.
- D Handley, B K Ghosh. Subcellular distribution of marker enzymes in cells of a minute fungus, Fusarium sp. 100-3. J Bacteriol.
- E T Brake , P C Will, J S Cook (2009). Characterization of HeLa 5′-nucleotidase: a stable plasma membrane marker. Membr Biochem. 1978;2(1):17-46.
- M. K. Johnson & V. P. Whittaker (1963). Lactate dehydrogenase as a cytoplasmic marker in brain. Biochem J, .88(3).
- Jean P.Dzoyem. VictorKuete. Jacobus N.Eloff. 2014. 23 – Biochemical Parameters in Toxicological Studies in Africa: Significance, Principle of Methods, Data Interpretation, and Use in Plant Screenings. Toxicological Survey of African Medicinal Plants. 659-715.
- J W Greenawalt, C Schnaitman (1970). An appraisal of the use of monoamine oxidase as an enzyme marker for the outer membrane of rat liver mitochondria. J Cell Biol. 46(1):173-9.
- Eigentler A, Draxl A, Gnaiger E (2020). Laboratory Protocol: Citrate synthase a mitochondrial marker enzyme . Oroboros