Proteasome Definition
The proteasome is a protein complex that plays a crucial role in maintaining the cellular homeostasis by degrading the misfolded or damaged protein molecules.
Proteasome complex is involved in following activities
- The proteasome is involved in MHC class I-restricted antigen processing of ER-targeted proteins and plays a major role in the processing of cytosolic proteins.
- Proteasome complex protects cancer cells from both tumor suppressor proteins and cytotoxic protein degradation process.
- Involved in the regulation of the cell cycle by reducing the availability of cyclin proteins and other key factors required for cell division.
- Proteasomal mechanism is linked to apoptosis, which is a form of programmed cell death that eliminates abnormal cells without releasing harmful materials into the surroundings. In general, proteasomes degrade specific proteins during apoptosis; however, in few cases, some components of the proteasome system are also degraded by apoptotic caspases.
- Involved in both biosynthesis and function of the cellular organelles including mitochondria, chloroplast, ER, Golgi, lysosomes, centrosomes, and ribosomes.
- Proteasome assists cell division by degrading the anaphase inhibitors.
- Regulates DNA transcription and DNA repair.
- Plays a central role in cell differentiation and development.
- Involved in neural and muscular degeneration.
- Regulates dynamic cellular mechanisms such as endocytosis, proteolysis, and immune signaling.
- Involved in morphogenesis of neural networks.
- Involved in the inflammation and immune responses.
- This protein plays a major role in the modulation of cell surface receptors, ion channels, and secretory pathways.
- Regulates bone remodeling mechanisms.
- Involved in the inhibition of osteoclastogenesis by degrading the RANKL; thus directly inhibitis the osteoclastic bone resorption.
- Ubiquitin-proteasome machinery can also regulate osteoblast bone formation by inhibiting the osteoblastogenesis.
- Responses to stress and extracellular modulators.
- Involved in the ribosome biogenesis.
- Regulates viral infection by degrading the protein coat of the virus.
1. Role of proteasome in the eukaryotic cell
- Regulation of protein hydrolysis and maintance of healthy proteins in the eukaryotic cells are crucial for cell viability through the elimination of more toxic, misfolded, damaged, or/and unwanted proteins before they accumulate to reach toxic levels.
- This important molecular proteolytic mechanism plays a pivotal role in the regulation of the cell cycle by controlling the concentrations of cyclin and other important proteins in a time-dependent manner.
- It also plays a role in the immune response by generating the antigenic peptides.
- In recent studies, surprisingly, proteasomal complex machinery is considered as a major drug target to control wide category of cancers.
2. Discovery of proteasome complex
Avram Hershko and his graduate student, Aaron Ciechanover collaboration with other scientists, Irwin Rose were discovered a ubiquitin-dependent protein degradation in the late 1970s and early 1980s and they were awarded Nobel Prize jointly in Chemistry in 2004 for the discovery of Ubiquitination.
3. Proteasome structure
- Proteasomes are multisubunit protease enzymes and cylindrical in shape.
- Proteasomes mediated ubiquitination process has been found in both prokaryotes and eukaryotes.
- In eukaryotes, they are found in both cytosol and nucleus.
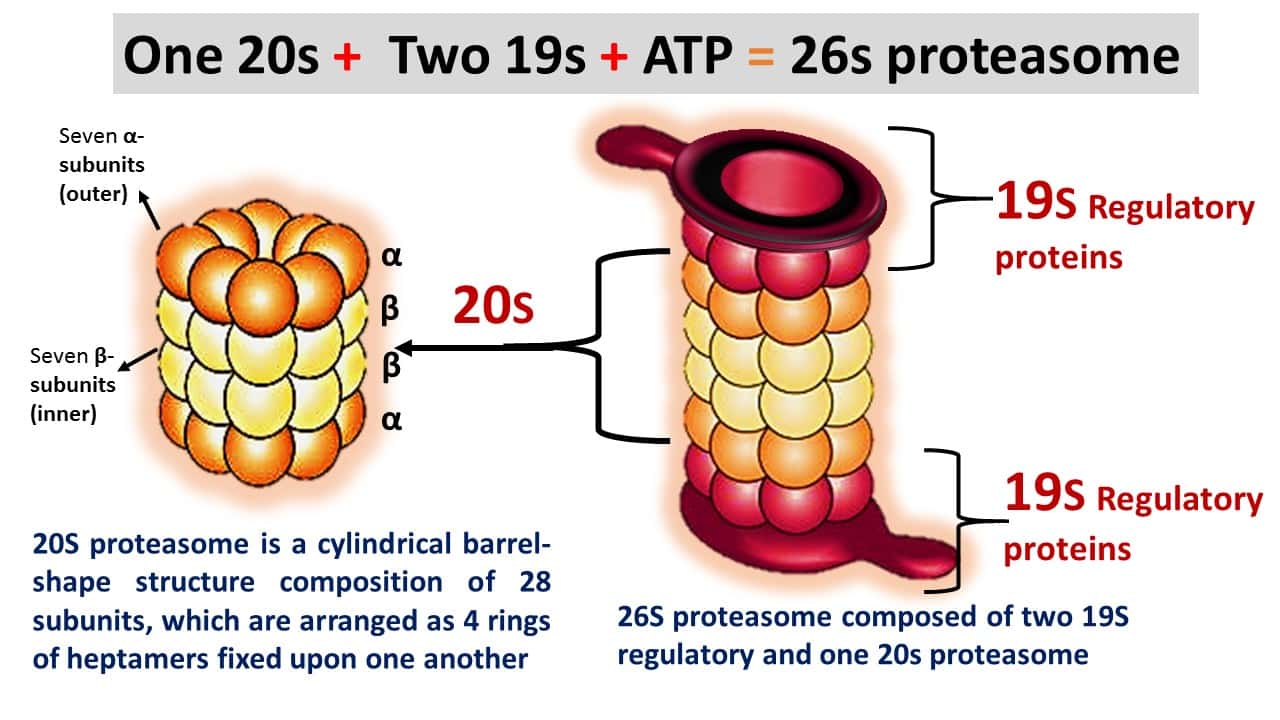
- Proteasomes of eukaryotes are two types, the 20S and 26S proteasomes.
- The 20S proteasome is a cylindrical barrel-shaped structure comprised of 28 subunits, which are arranged as four rings of heptamers, which are fixed upon one another.
- These all subunits belong to the same superfamily of proteins, however, which are grouped into two families, alpha and beta.
- In eukaryotes, 26S type proteasome is formed by the combination of two 19S regulatory proteins, which is an ATP-required process (Figure 1).
4. Proteasome function
- The 26S proteasome digests proteins by using cellular energy; however, these proteasomes can not degrade cellular proteins without proteolytic signals, which are generated from the ubiquitination process.
- Ubiquitination-mediated proteolysis is a regulatory process, that involves various regulatory proteins. The ubiquitination mechanism produces polyubiquitinated proteins (susceptible proteins), which will be degraded by the proteasome complex.
(i) The function of 20S proteasome core
- The 20S core particle of the proteasome complex is a molecular machine that plays an important role in the degradation of misfolded or unwanted protein substrates, which is a non-lysosomal protein degradation, thereby ensuring protein homeostasis in eukaryotic cells.
(ii) The function of 19S regulatory units
- 19S regulatory unit (gate) has an ATPase activity, which regulates the function of proteasome via a gating mechanism.
- Gating mechanism controls sizes of the pores at the top and bottom ends of the symmetric proteasome barrel and restricts access to catalytic sites sequestered in the lumen of the structure.
- The unit is mainly involved in three functions. First, the unit recognizes and binds specifically to polyubiquitin chains, thereby ensuring that only ubiquitinated protein to be degraded.
- The other function include 19S unit removes intact molecules from the fated protein so that they can be reused.
- The third function; the 19S unit unfolds fated proteins and sends them into catalytic lumen or core.
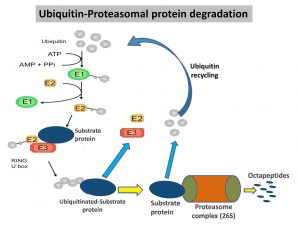
5. Ubiquitin-Proteasomal protein degradation process
- The reaction starts with the attachment of ubiquitin proteins to the damaged protein.
- However, it happens in three steps. In the first step, the terminal carboxyl group of the ubiquitin interacts with ubiquitin-activating enzyme (E1) by thioester bonding using an energy released from ATP hydrolysis.
- In the second step, the activated ubiquitin protein is subsequently transferred to a sulfhydryl group of the ubiquitin-
conjugating enzyme (E2). - In the final step, ubiquitin-ligase (E3) catalyzes the separation of attached ubiquitin molecules from E2 and transfers them to the target protein (specific substrate proteins).
6. Proteasome as a target for cancer treatment
- The continuous proliferation of cancer cells requires both immediate turnover of expired or misfolded proteins and the efficient degradation of tumor suppressor proteins and pro-apoptotic factors, e.g., p53 protein.
- Because this immediate turnover provides free amino acids to reuse for the synthesis of new proteins in cancer cells.
- Moreover, efficient degradation of tumor suppressors and pro-apoptotic proteins protects the cancer cells from self-degradation.
- Due to this advanced feature, cancer cells are more powerful than normal cells to control the cytotoxic effect, which is formed by the accumulation of unwanted proteins.
7. Proteasome inhibition by different inhibitors
- In earlier studies, scientists have recognized numerous small proteasome complex inhibitory drugs.
- Especially, the inhibitors are related to the peptide aldehyde group that inhibits proteasome function reversibly and they have been explored extensively in cancer research.
- While they also inhibit the function of cellular proteases.
- Since they have the ability to control protease activity, they have been used as potential drugs against different cancers.
Beta-lactone: Beta-lactone can inhibit the proteasome function by binding irreversibly to the amino acid, threonine found in the active-site of the β subunits in the 20s proteasome. But they are not completely proteasome-specific inhibitors.
Epoxyketone class: Carfilzomib and Oprozomib are epoxyketone classes of proteasome inhibitors, which are more specific to the proteasome.
Dipeptidyl boronic acid class: The bortezomib is a dipeptidyl boronic acid class proteasome inhibitor that can inhibit the proteasome function. The recent studies stated that the application of bortezomib during bone formation induces osteoblast differentiation by protecting the differentiation-related transcription factors from proteasome-mediated protein degradation.
Disulfiram: Disulfiram and its analog pyrrolidine dithiocarbamate (PDTC) have been recognized as partial non-competitive proteasome inhibitors, which can bind slowly to 20S proteasome and inhibit its catalytic activity. Hence, these are used against chronic lymphocytic leukemia.
Marizomib: Marizomib (salinosporamide A) has been used against multiple myeloma. The drug can inhibit all three subunits of the proteasome complex.
8. Other proteasome inhibitor drugs
Delanzomib is also used against wide category of cancers.
Epoxomicin is a naturally occurring selective inhibitor drug for protein degradation.
Epigallocatechin-3-gallate has also been proposed as a potent inhibitor.
Beta-hydroxy beta-methyl butyrate is a proteasome inhibitor and it is used to improve human skeletal muscle building.
References
- Banaszynski et al., “A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules,” Cell, 126:995-1004, 2006.
- Garrett IR1, Chen D, Gutierrez G, Zhao M, Escobedo A, Rossini G, Harris SE, Gallwitz W, Kim KB, Hu S, Crews CM, Mundy GR. Selective inhibitors of the osteoblast proteasome stimulate bone formation in vivo and in vitro. J Clin Invest. 2003 Jun;111(11):1771-82.
- Gustafson et al., “Small molecule mediated degradation of the androgen receptor through hydrophobic tagging,” Angew Chem Int Ed Engl, 54:9659-62, 2015.
- Uyama M1, Sato MM, Kawanami M, Tamura M. Regulation of osteoblastic differentiation by the proteasome inhibitor bortezomib.Genes Cells. 2012 Jul;17(7):548-58. doi: 10.1111/j.1365-2443.2012.01611.x. Epub 2012 Jun 15.
- Bondeson et al., “Catalytic in vivo protein knockdown by small-molecule PROTACs,” Nat Chem Biol, 11:611-17, 2015.
- Jeanmarie Verchot. Plant Virus Infection and the Ubiquitin-Proteasome Machinery: Arms Race along the Endoplasmic Reticulum. Viruses. 2016 Nov; 8(11):
- Schneekloth et al., “Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics,” Bioorg Med Chem Lett, 18:5904-08, 2008.
- Winter et al., “Phthalimide conjugation as a strategy for in vivo target protein degradation,” Science, 348:1376-81, 2015.
- Lövborg H, Oberg F, Rickardson L, Gullbo J, Nygren P, Larsson R. Inhibition of proteasome activity, nuclear factor-KappaB translocation and cell survival by the antialcoholism drug disulfiram. Int J Cancer. 2006 Mar 15;118(6): 1577-80.
- Raina et al., “PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer,” PNAS, 113:7124-29, 2016.
- Wójcik C. Regulation of apoptosis by the ubiquitin and proteasome pathway. J Cell Mol Med. 2002 Jan-Mar;6(1): 25-48.
- Buckley et al., “HaloPROTACS: Use of small molecule PROTACs to Induce degradation of HaloTag fusion proteins” ACS Chem Biol, 10:1831-37, 2015.
- Orlowski RZ. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999 Apr;6(4): 303-13.
- Hines et al., “Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs” PNAS, 110:8942-47, 2013.
- Hasinoff BB, Patel D. Disulfiram is a slow-binding partial non-competitive inhibitor of 20S proteasome activity. Arch Biochem Biophys. 2017 Nov 1;633: 23-28. doi: 10.1016/j.abb.2017.09.003. Epub 2017 Sep 5.